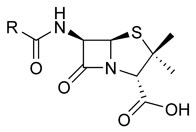
 Penicillin core
Penicillin core
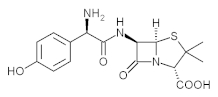 Amoxicillin
Amoxicillin
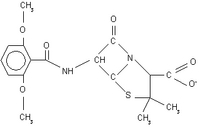 Methicillin
Methicillin
 Penicillin core
Penicillin core
 Amoxicillin
Amoxicillin
 Methicillin
Methicillin
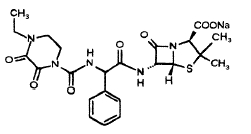 piperacillin
piperacillin
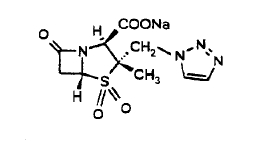 tazobactam
tazobactam
One solution is tazobactam, which is marketed in combination with piperacillin, binds in the active site of beta-lactamase preventing the beta-lactamase from destroying the pipericillin. From the chemical structure it is clear that the molecule resembles closely the penicillin type antibiotics and has a good chance at binding int the active site of the betalactmases. It appears that the mechanism of inhibition is the chemical bonding of a fragment to an active site serine, leaving the active site permanently disabled. The chemical binding of the fragment occurs as part of the degrading of the tazobactam molecule.
This tutorial is an introduction to setting up and starting simulations of protein-ligand systems with a nonstandard ligand. The aim of the project is to model the docked ligand to get structural and thermodynamic data about the bound ligand.